How we work within food microbiology – important topics … and the techniques and conceptual approaches that make it happen.
Molecular methodologies dominate practical aspects of modern biology – food, environmental and agricultural microbiology is no exception. However, it is more important for analytical methodologies in these areas of microbiology to focus on accuracy and precision rather than speed. In other areas of food microbiology, such as food safety investigations during disease outbreaks, where rapid results are especially important, rapid and sometimes real-time methods are vital. However, speed is relatively unimportant when detecting and identifying microorganisms in the context of food fermentations, environmental microbiology studies and agricultural microbiology. Thus, time to result is relatively unimportant, whereas getting as close as possible to the correct result is paramount, even if the “correct result” is an unknown bacterial species. Sometimes, obtaining any result may not be possible at all, which is one of the challenges.
Unlike other areas of microbiology, like food safety outbreak investigations, many a time, work in these areas has more of a focus on investigation and detection of non-standard or rare microorganisms. Such investigations could very well involve novel microorganisms, completely new to science. Therefore, by and large, a different approach is required, compared to those fields of microbiology which routinely involve investigation of typical, known and expected microorganisms. The idea ‘you find what you look for’ is so true in these particular microbiology analyses, where, in the case of environmental samples, very few microorganisms may be culturable, despite being metabolically active. Consequently, what doesn’t work that well in environmental microbiology investigations are comparison of results to known databases or using known PCR primers. As a result, investigative protocols based on 16s rRNA tend to work best (Benlloch et al., 1995). There will still be some limitations if a PCR methodology is employed, but independent testing regimes exist (Rosselli et al., 2016), which therefore do away with the limitation of the use of primers to known gene sequences.
- Molecular and spectroscopic approaches to analytical microbiology
Prior to the era of routine molecular biology, traditional cultural and biochemical methods, relying on genetic differences manifesting as observable phenotypic differences, were the mainstay of bacterial detection and identification, in food microbiology and indeed other areas of microbiology. However, since 1995 when the first bacterial genome was sequenced, molecular genetics, in particular of whole genomes, has moved into the area of semi-routine. Previously limited by technical capabilities, the shortcomings of combining numerous short DNA sequences were largely overcome, at least in a practical sense in certain applications by pulsed field gel electrophoresis (PFGE). However, this has been rapidly superseded by whole genome sequencing (WGS), used increasingly for routine foodborne disease outbreak investigations of an epidemiological nature. This allows for unprecedented levels of precision in strain distinction and hence for determining relatedness, allowing for previously unmatched levels of understanding in the development and movement of bacterial pathogens.

In addition, the interface of biology, chemistry, physics and mathematics as a combination in various areas of applied microbiology, allows for the development of practical approaches to solving real-world problems in food microbiology that have never been possible. For example, while the use of near infrared spectroscopy (NIRS) isn’t new in the context of food quality analysis, it is now being refined and extended into a new level of precision for pathogenic bacterial detection in food products. This, and other approaches, such as electrochemical-based sensor technology for detection of spoilage bacteria within food products offers quick and precise analysis that isn’t invasive. Overall, the field of molecular microbiology is more prominent than ever, with various omics techniques, in particular genomics, proteomics and metabolomics, becoming an increasing indispensable part of modern microbiology. As in all areas of biology, going (or gone!) are the days of purely phenotypic characterisation as being sufficient. Today demands a greater understanding, to levels never before seen, as we delve deep into cell biology and genetics, unlocking secrets with increasing capabilities due to contributions from diverse fields of chemistry, physics and mathematics, that combine, to propel analytical microbiology to a whole new era in the coming years.
- Progress on approaches to grow microorganisms that cannot be grown as yet,
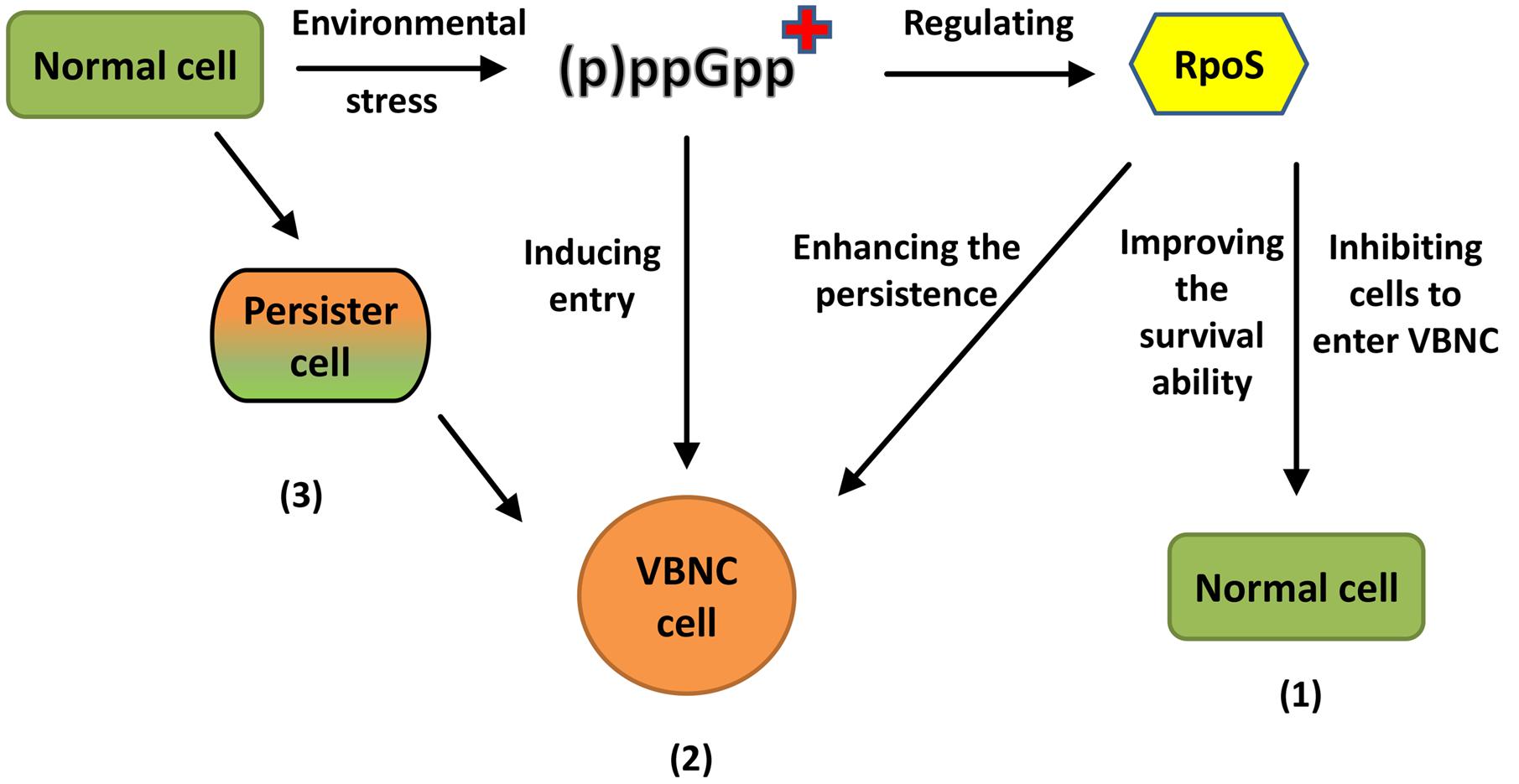
The topic of this section can be related to either those bacterial cells that have never been cultured, and it is understood that an estimated 99% of all species of Archaea and Bacteria are yet to be cultured, or in the so-called viable but non-culturable (VBNC) state, a concept developed some 40 years ago by Xu et al. (1982) to describe the survival mechanism state where normally culturable bacterial cells that are metabolically active, enter into a state of being unable to be cultured. This would essentially result in false negative readings in microbiologically food analysis, where the metabolically active bacterial cells are in fact present, but although unable to grow and form colonies on artificial media, may certainly still be virulent and remain in a state of pathogenicity. Thus, Fakruddin et al. (2013) states that negative microbiological test results from food products cannot necessarily be considered from pathogens. A key feature of the preceding section is the fact that molecular level and omics techniques are contributing to bring forth elements of discovery previously unheard of, and thus enabling their incorporation into routine commercial, industrial and/or diagnostic practice. This too, is a key feature here, where DNA analysis techniques are likely to offer improved opportunities for exploring the largely unexplored microbial world in the context of food microbiology. This could very well provide some breakthrough moments in industrial and/or public health challenges and furthermore, may offer new opportunities to be developed, in potentially unknown ways, from the identification, genome sequence of and industrial/research use of potentially undiscovered microorganisms. A practical consideration is how to survey and collect microorganisms from a whole range of extreme environments on our planet. There were great limitations on advancements in this field prior to the widespread availability of molecular analysis tools. Despite the benefits of a molecular-based approach, Rampelotto (2013) states that the modern techniques of molecular biology, when used in combination with traditional cultural methods, will deliver the best approach.
References
Benlloch, S., Martínez-Murcia, A.J. and Rodríguez-Valera, F., 1995. Sequencing of bacterial and archaeal 16S rRNA genes directly amplified from a hypersaline environment. Systematic and Applied Microbiology 18:574-581. https://doi.org/10.1016/S0723-2020(11)80418-2
Fakruddin, M.d., Mannan, K.S.B. and Andrews, S., 2013. Viable but nonculturable bacteria: Food safety and public health perspective. International Scholarly Research Notices 2013. https://doi.org/10.1155/2013/703813
Rosselli, R., Romoli, O., Vitulo, N., Vezzi, A., Campanaro, S., de Pascale, F., Schiavon, R., Tiarca, M., Poletto, F., Concheri, G., Valle, G. and Squartini, A., 2016. Direct 16S rRNA-seq from bacterial communities: a PCR-independent approach to simultaneously assess microbial diversity and functional activity potential of each taxon. Scientific Reports 6. https://doi.org/10.1038/srep32165
Xu, H.S., Roberts, N., Singleton, F.L., Attwell, R.W., Grimes, D.J., and Colwell, R.R., 1982. Survival and viability of nonculturable Escherichia coli and Vibrio cholerae in the estuarine and marine environment. Microbial Ecology 8:313-323.